Scientists at the Keck School of Medicine of USC have discovered that a protein known as PICALM regulates removal of toxic plaques from the brain, which could be a potential therapeutic target for the treatment of Alzheimer’s disease.
In a study that appears in the May 25 edition of the scientific journal Nature Neuroscience, researchers identify this new role for PICALM, which is a known genetic risk factor for Alzheimer’s disease.
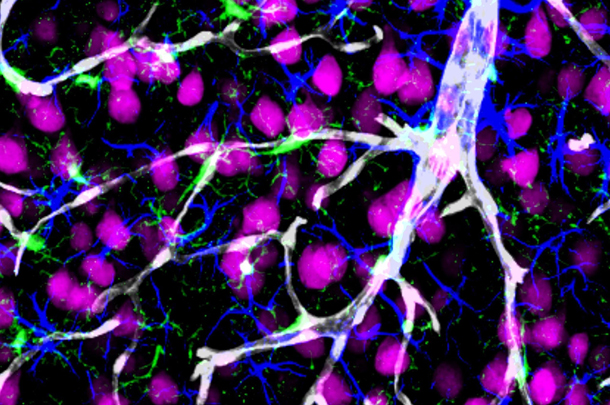
Alzheimer’s is the most common type of dementia, characterized by the loss of memory and other mental abilities linked to an accumulation of amyloid-beta and other toxic compounds in the brain. The study found that a deficiency in PICALM in cerebral blood vessels and in PICALM-related gene variants associated with increased risk for Alzheimer’s, disable amyloid-beta from being cleared out of the brain across a region known as the blood-brain barrier.
“There have been many new genes discovered to be associated with Alzheimer’s disease, but the biology of these genes are poorly understood,” said the study’s principal investigator Berislav Zlokovic, M.D., Ph.D., director of the Zilkha Neurogenetic Institute and holder of the Mary Hayley and Selim Zilkha chair for Alzheimer’s Disease research at the Keck School of Medicine of USC. “Our new study shows that a deficiency in PICALM in blood vessels and its variants associated with increased risk for the disease inactivate amyloid-beta clearance from the brain, leading to its accumulation and cognitive impairment. This new study provides fundamental new information about PICALM and brings to light novel potential therapeutic targets for increasing amyloid-beta clearance in Alzheimer’s disease.”
Autopsies from Alzheimer’s disease patients and recent research in experimental models have shown the importance of brain blood vessels in the disease’s initiation and progression. For more than two decades, Zlokovic and his research team have studied the cellular and molecular mechanisms of brain blood vessels that maintain normal cognition with hopes of developing new treatments for Alzheimer’s and other neurodegenerative diseases. One focus of their lab at the Zilkha Neurogenetic Institute is on PICALM, or phosphatidylinositol binding clathrin assembly protein, which in humans is encoded by the PICALM gene.
By performing a neuropathological study in humans with Alzheimer’s and using transgenic animals to model the disease, the group found that low levels of PICALM in brain endothelial cells lead to amyloid-beta accumulation in the brain. Genetic variants associated with the PICALM gene have been shown to increase the risk of Alzheimer’s disease. The researchers also generated human endothelial cells from induced pluripotent stem cells to examine the consequences of a known PICALM variant associated with increased risk for Alzheimer’s; they found that this genetic alteration disrupted amyloid-beta clearance by cerebral blood vessels.
These new findings have prompted Zlokovic to address new questions about the role of PICALM in Alzheimer’s disease. Future studies will explore how genetic flaws in the PICALM gene influence its expression levels and clearance function at the blood-brain barrier and the general health of cerebral blood vessels. The team also will work on developing therapeutic strategies, including gene therapy, and screening for new drugs to overcome PICALM deficiency.
Co-authors on the study from USC include Zhen Zhao, Abhay Sagare, Qingyi Ma, Matthew Halliday, Pan Kong, Kassandra Kisler, Ethan Winkler, Anita Ramanathan, Nelly Chuqui Owens, Sanket Rege, Gabriel Si, Ashim Ahuja, Carol Miller, Tohru Sugawara and Justin Ichida.
The study was funded in part by the National Institutes of Health (R37NS34467, R37AG23084, R01AG039452, R01AG035355 , R01AG027924, R00NS07743), Cure for Alzheimer Fund, American Cancer Society (RSG–13–379–01–LIB), Rainwater Charitable Foundation, Donald E. and Delia B. Baxter Foundation, and Daiichi Sankyo Foundation of Life Science. This study used sample ND10689 from the NINDS Cell Line Repository, as well as clinical data.
— Les Dunseith